A Deep Dive into Sequencing Technologies
Estimated Reading Time: 7 minutes
TL;DR
In our last issue, we provided a brief introduction to genomic sequencing but did not spend much time on the actual technology. In today’s issue, we take a deeper look at the science underlying modern sequencing. We start first with a few announcements below. Our next issue will be released in roughly 2 weeks.
Differentiable Physics: A Position Piece
We’ve been hard at work on a number of internal projects here at Deep Forest Sciences and I’m excited to finally be able to share one of them publicly. We’ve just released our new perspective, Differentiable Physics: A Position Piece, written in partnership with Venkat Viswanathan and Dilip Krishnamurthy at CMU, out onto Arxiv, the scientific preprint server.
We believe differentiable physics, the use of differentiable programming for physical systems, is a powerful new tool for understanding physical systems, one which will have broad applicability in both scientific and industrial domains. We also emphasize the new role of “scientific foundation models,” broadly relevant scientific algorithms trained on large datasets.
Get in touch if you’d like to partner with us to apply differentiable physics or scientific foundation models to hard real-world problems! (bharath@deepforestsci.com)
DeepChem’s First Online User Group Meeting
DeepChem is the core open source technology we develop at Deep Forest Sciences. If you’re interested to learn more about DeepChem, please attend our first online user group meeting. DeepChem’s Google Summer of Code interns will present on their work for the summer, adding support for partial differential equations, small molecule retrosynthesis, protein language modeling, and improved molecular machine learning to DeepChem.
Polymerase Chain Reaction
The Polymerase Chain Reaction (PCR) is one of the most foundational genomic technologies. It allows for exponential amplification of genomic sequences as shown in the figure below. Many sequencing technologies use PCR to amplify small amounts of DNA for detection.

Thermal cycling (repeatedly heating and cooling reactants) enables PCR by favoring different temperature dependent reactions (DNA melting vs enzymatic DNA replication) at different temperatures. At the first step, DNA is heated until it denatures (that is, separates into two separate strands of single-stranded DNA). The DNA is then cooled, and the two single stranded DNA strands are used as templates by the DNA polymerase enzyme to assemble two strands of double stranded DNA. Note that at the end of this process, the original DNA strand has been doubled! By repeating this process, the original DNA strand can be exponentially amplified.
PCR is a fascinating procedure since it exploits the discrete combinatorial structure of DNA. There exists no natural or synthetic procedure to duplicate an arbitrary small molecule for example. I’d characterize PCR as a type of “physical algorithm;” that is, an algorithmic procedure that operates on a real physical substrate. The field of DNA computing has explored the use of DNA as a framework for more general purpose computing. At present, the challenge is that while certain special-purpose algorithms can be solved very effectively on a DNA-based computer, there does not yet exist a procedure to solve general computing problems in programmatic fashion on a DNA substrate. DNA storage might prove to be a breakout use for DNA-based technologies though since it enables compact storage of very large datasets.
Sanger Sequencing
Sanger sequencing exploits PCR to sequence a DNA strand. A variant of PCR is executed at which duplication of the original strand cuts off at a random prefix of the original strand. That is, if the original strand was N nucleotides long, the duplication procedure will create duplicates of the length 1, 2, 3, …, N prefixes of the original DNA sequence. These prefixes are capped with dideoxynucleotides, ddNTPs, that terminate the action of DNA polymerase. Gel electrophoresis is used to separate these different prefixes by size. The sequence can be read directly off the gel manually, or alternatively by laser excitation/detection for rapid sequencing. A scaled-up form of Sanger sequencing was used to sequence the first human genome in 2001.

Nanopore Sequencing
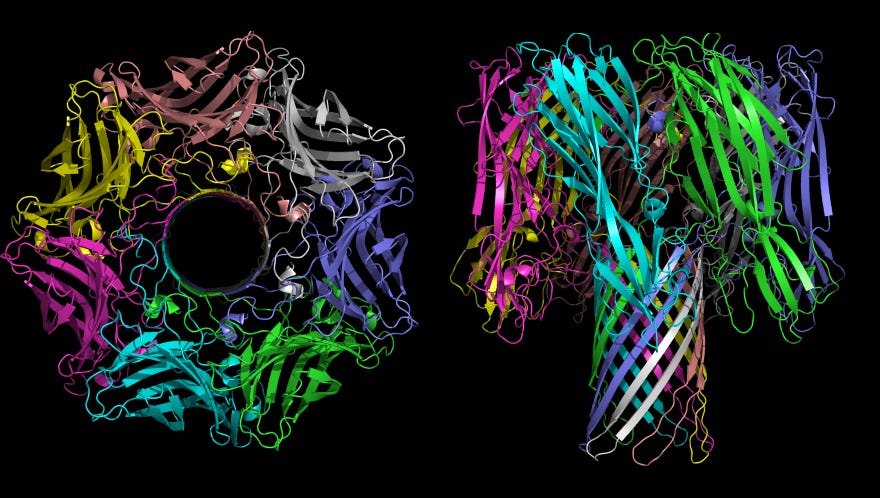
Nanopore sequencing bypasses the need for PCR amplification of a molecule by driving single stranded DNA through a protein nanopore embedded in a thin membrane (see figure below). Nanopore sequencing holds out the powerful promise of being able to do “long-reads” which don’t require fragmentation of the DNA molecule into multiple short segments. These technologies are still early, but have been greatly matured by the hard work of Oxford Nanopore.

Other Sequencing Technologies
A remarkable array of different technologies have been proposed for use in sequencing and we’ve only covered the very basics. Some prominent methods include microfluidics-based sequencing, semiconductor sequencing, mass spectrometry sequencing, and microscopy based sequencing.
Weekly News Roundup
https://www.wsj.com/articles/biden-administration-officials-try-to-soothe-france-over-australia-submarine-deal-11631821352: Dueling moves by the US and the PRC in the Pacific.
https://www.wsj.com/articles/u-s-steel-plans-new-u-s-mill-as-prices-surge-11631827557: US Steel is planning a new steel mill in the US.
https://www.quantamagazine.org/new-math-book-rescues-landmark-topology-proof-20210909/: A fascinating story about a quest to “rescue” an esoteric proof.
https://www.wsj.com/articles/taiwan-plans-to-bulk-up-military-budget-to-contend-with-chinese-pressure-11631787522: Taiwan is bulking up its defense budget.
Feedback and Comments
Please feel free to email me directly (bharath@deepforestsci.com) with your feedback and comments!
About
Deep Into the Forest is a newsletter by Deep Forest Sciences, Inc. We’re a deep tech R&D company specializing in the development of AI for deep tech applications. Partner with us to apply our foundational AI technologies to hard real-world problems. Get in touch with us at partnerships@deepforestsci.com!
Credits
Author: Bharath Ramsundar, Ph.D.
Editor: Sandya Subramanian, Ph.D.