Advancing Protein Sequencing Using Nanopores
Estimated Reading Time: 3 minutes
TL;DR
This week, we have a brief post discussing a recent article introducing a novel method for reading long protein strands at the single-amino acid level using nanopores and an unfoldase motor protein.
Advancing Protein Sequencing Using Nanopore Reads for Long Protein Strands
The ability to sequence proteins would enable us to deepen our understanding of proteomics and better understand the behavior of cells. Proteoform diversity is a term used to indicate the diversity in protein forms resulting from transcription, translation, and post-translational modifications (PTMs). Such changes play crucial roles in biological processes and disease states. Current protein sequencing methods, such as Edman degradation and mass spectrometry have limitations in sensing such modifications effectively. Improved sequencing technology could enable us to understand proteoform diversity and could potentially lead to improved therapeutics down the road.
Nanopore technology involves using nanometer-sized pores within an insulating membrane to drive ionic current flow when individual analyte molecules pass through the pore. While initially developed for DNA sequencing, nanopore methods could hold significant potential for protein analysis by enabling discrimination of peptides and proteins, improving measurement of protein-protein interactions, and detecting PTMs like phosphorylation and glycosylation. However, sequencing entire protein strands with nanopores has been challenging thus far due to aspects of protein structures such as neutral backbones and varying charge states of side chains. (It is also worth noting that a number of non-nanopore protein sequencing techniques such as single-molecule fluorescence labeling and affinity-based approaches have been proposed in the literature, but these methods have their own tradeoffs.)
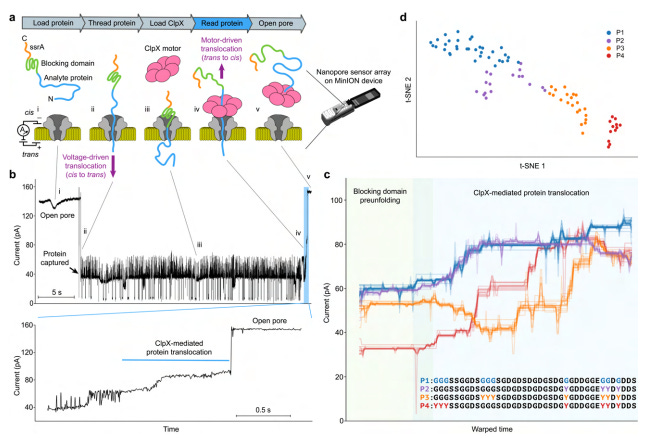
This article presents a novel approach involving electrophoresis and ClpX unfoldase to sequence long protein strands at the single-amino acid level. The method leverages a commercial MinION nanopore sequencer combined with unfoldase to enable the detection of single amino acid substitutions and PTMs in long protein sequences. A machine-learned model is used to perform “aminocalling” and transform raw readouts into protein sequences. Accuracy is still fairly low, but subsequent research will likely help improve performance. The paper also develops a biophysical model to simulate nanopore readouts directly from amino acid sequences, and demonstrates the potential for single-molecule protein identification.
This work is mostly a proof-of-concept for now, and several technical advances will be needed for nanopore protein sequencing to become a robust technique. However, there is clear potential for commercial protein nanopore sequences to become a powerful new biological tool in the years to come.
Interesting Links from Around the Web
https://www.quantamagazine.org/biophysicists-uncover-powerful-symmetries-in-living-tissue-20231025/: A cool write-up about improved biophysical models for tissues
https://www.nature.com/articles/d41586-023-03317-7: The FDA is set to examine the first CRISPR based therapy
https://www.nature.com/articles/s41586-023-06602-7: A proof-of-concept particle accelerator on a chip.
Feedback and Comments
Please feel free to email me directly (bharath@deepforestsci.com) with your feedback and comments!
About
Deep Into the Forest is a newsletter by Deep Forest Sciences, Inc. We’re a deep tech R&D company building Chiron, an AI-powered scientific discovery engine for the biotech/pharma industries. Deep Forest Sciences leads the development of the open source DeepChem ecosystem. Partner with us to apply our foundational AI technologies to hard real-world problems in drug discovery. Get in touch with us at partnerships@deepforestsci.com!
Credits
Author: Bharath Ramsundar, Ph.D.
Editor: Sandya Subramanian, Ph.D.
Research and Writing: Rida Irfan