Enhancing Virtual Screening with Electron Density Data
Estimated Reading Time: 3 minutes
TL;DR
This week, we cover a recent article discussing the use of electron densities to improve virtual screening methods.
Using Experimental Electron Densities to Enrich Active Compounds in Virtual Screening
Although structured based virtual screening methods are widely used, their practical accuracy remains limited. Practitioners are often happy if a few predicted hits confirm in practice. This article introduces a new technique for using experimental electron densities to improve virtual screening accuracy. Such electron densities can be obtained from X-ray crystallography or Cryo-EM experiments on the protein in question.
The authors propose a multi-resolution Experimental ED-based grid matching score (ExptGMS), a scoring method based on experimental electron density (ED) maps. ED maps represent time-averaged distributions of molecules in crystalline structures and contain dynamic information about ligands and solvents in the binding pockets. Such information is often overlooked in traditional structure-based virtual screening methods which operate directly from processed structures.

ExptGMS consists of an experimental ED based grid and a scoring function. The grid assigns values based on the ED intensity at specific positions within and around the binding pockets. The scoring function evaluates ligand conformers based on how well they match with the grid, rewarding ligand atoms occupying grid points with strong ED intensity and penalizing both atoms occupying regions lacking ED information and grid points with strong ED but no corresponding ligand atom. The authors train a gradient-boosted decision tree (GBDT) on extracted structural features combined with ExptGMS scores as descriptors to produce a binary prediction of binding. Models are trained on a collection of targets and tested for accuracy on held-out targets.
ExptGMS, coupled with the GBDT model, appears to provide slight boosts to predictive power as measured on the DUD-E dataset. The paper runs a virtual screen for Covid-19 3CLpro inhibitors using ExptGMS combined with docking scores and validated in a downstream assay. As a control, compounds selected only with docking were also tested in the downstream assay. The authors claim the combined method yields three times more experimentally active compounds for 3CLpro than docking scores alone. If this claimed result holds for other targets, it would be a powerful argument for making more systematic use of electron densities in virtual screens.
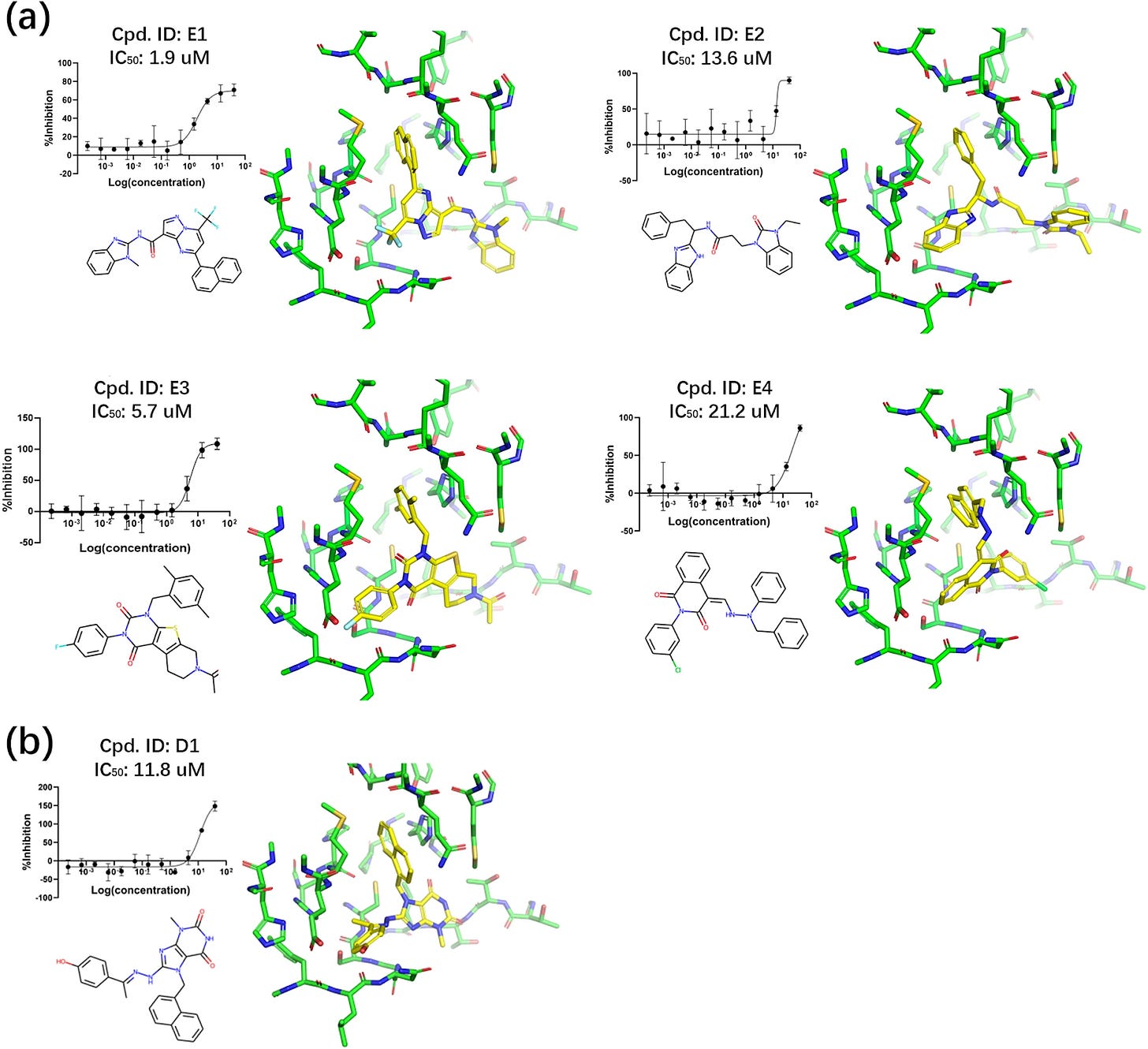
ExptGMS heavily depends on the accuracy of the initial binding pose from a docking method, which potentially limits its effectiveness if the pose is incorrect. Moreover, it also relies on experimental ED data, which may not always be accessible, particularly in cases involving allosteric pockets. The authors indicate it may be possible to use a computational method to estimate the electron density in this case, but suggest it as a direction for future research. The authors haven’t open sourced the code for their study, so while the technique is interesting, it may take some time until other groups can replicate and verify the results.
Interesting Links from Around the Web
https://arxiv.org/abs/2310.03011: This extensive wiki-like write-up of the quantum algorithms and their applications is a really powerful overview of the field. Real world applications of quantum computing are on the horizon.
https://www.nature.com/articles/s41586-023-06563-x: Large scale proteomics is coming of age.
https://www.science.org/content/article/laser-mapping-reveals-hidden-structures-in-amazon-hints-thousands-more: Lidar is revealing that there may be thousands of human earthwork structures through the Amazon.
Feedback and Comments
Please feel free to email me directly (bharath@deepforestsci.com) with your feedback and comments!
About
Deep Into the Forest is a newsletter by Deep Forest Sciences, Inc. We’re a deep tech R&D company building Chiron, an AI-powered scientific discovery engine for the biotech/pharma industries. Deep Forest Sciences leads the development of the open source DeepChem ecosystem. Partner with us to apply our foundational AI technologies to hard real-world problems in drug discovery. Get in touch with us at partnerships@deepforestsci.com!
Credits
Author: Bharath Ramsundar, Ph.D.
Editor: Sandya Subramanian, Ph.D.
Research and Writing: Rida Irfan

