Supporting Open Source and Extending AlphaFold to Protein-Ligand Interactions
Estimated Reading Time: 3 minutes
TL;DR
This week, we discuss the importance of supporting open source and cover a recent article from DeepMind that discusses their latest (unpublished) improvements to AlphaFold.
Protecting Open Source AI
If you are an advocate or supporter of open source and open source AI, I strongly encourage you to consider signing the open source AI letter (https://open.mozilla.org/letter/). Misguided AGI fears are threatening to harm societally useful open source efforts.
AlphaFold-latest: AlphaFold for Protein-Ligand Modeling
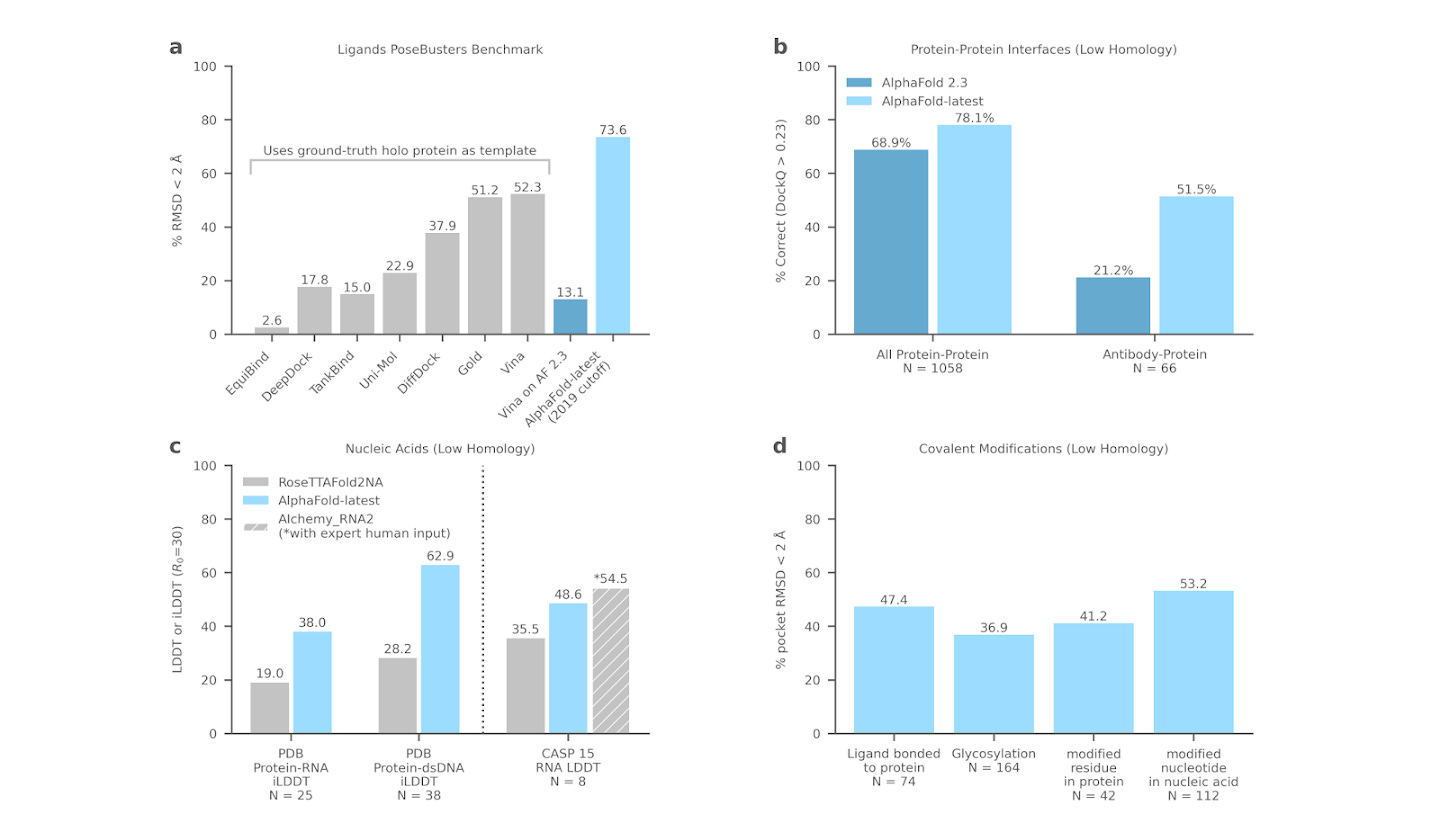
DeepMind’s article discusses progress made in the development of its next generation AlphaFold system, designed to predict joint protein-ligand/protein-nucleic acid structures. DeepMind has been working over the last few years to extend AlphaFold’s capabilities to predict the 3D structures of various biologically relevant molecules, not limited to just proteins.
This latest iteration of AlphaFold appears to generate highly accurate predictions for a wide range of molecules, including ligands, proteins, nucleic acids. It can also handle post-translational modifications of proteins. These molecular structures collectively play a vital role in understanding biological processes but have been challenging to model accurately with computational methods.
The report highlights what looks like strong performance in predicting protein-ligand structures. DeepMind claims AlphaFold-latest considerably outperforms traditional docking methods, like AutoDock Vina, and achieves a 73.6% success rate at predicting high fidelity protein-ligand structures, compared with a 37.9% success rate for DiffDock. It also appears that AlphaFold-latest does a strong job of predicting ligand-protein and nucleic acid-protein interactions for novel proteins without requiring a reference structure and appears to improve state-of-art protein-ligand structure prediction. DeepMind claims their model can represent the inherent flexibility of proteins and nucleic acids as they interact with other molecules, but does not reveal many algorithmic details about how this feat is achieved.
DeepMind tests AlphaFold-latest on three therapeutically relevant targets: PORCN, an integral membrane protein; KRASγ, an important cancer target; and PI5P4K, phosphoinositide kinases. Looking at the results, it does indeed appear like DeepMind has a practically useful system, but it’s hard to know how the tool would perform on other proteins or systems of interest.
DeepMind’s results appear impressive and show strong claimed improvements over both DiffDock and Autodock Vina. However, this note contains no details about the architecture and training methodology of their system, in line with the unfortunate new standard of not open-sourcing commercially useful AI. For this reason, it’s perhaps more useful to think of the current work as a technical whitepaper advertising DeepMind’s/Isomorphic’s new offering than as a rigorous scientific manuscript. DeepMind themselves only call their write-up a “progress report.” I hope DeepMind eventually publishes details of their method, and I also hope to see academic and open source groups replicate some of the claimed results and elucidate the underlying scientific principles.
Interesting Links from Around the Web
.https://spectrum.ieee.org/diabetes: An interesting new wound-healing magnetic gel.
Feedback and Comments
Please feel free to email me directly (bharath@deepforestsci.com) with your feedback and comments!
About
Deep Into the Forest is a newsletter by Deep Forest Sciences, Inc. We’re a deep tech R&D company building Chiron, an AI-powered scientific discovery engine for the biotech/pharma industries. Deep Forest Sciences leads the development of the open source DeepChem ecosystem. Partner with us to apply our foundational AI technologies to hard real-world problems in drug discovery. Get in touch with us at partnerships@deepforestsci.com!
Credits
Author: Bharath Ramsundar, Ph.D.
Editor: Sandya Subramanian, Ph.D.
Research and Writing: Rida Irfan